OpenGeneMed Licensing Agreement
This software is free for non-commercial use. For technical issues, please contact first author (Alida Palmisano).
All users must agree to the following conditions:
- Please reference Palmisano et al. 2016 paper for all studies using OpenGeneMed.
- The package or any of its components must not be distributed to others without prior approval of the authors. The package is offered without support; the user must not hold the National Cancer Institute liable for any damages resulting from the use of OpenGeneMed.
- Download OpenGeneMed version 1.0 (released Dec. 2015)
Important: the Virtual Machine file has been created and fully tested with Virtual Box, version 4.3. This version can be downloaded free from the Virtual Box wiki webpage. Following versions of Virtual Box experienced issues with network connectivity independent from OpenGeneMed. Once your local VM is running, if you have issues connecting with OpenGeneMed, please verify that you are using the right version of Virtual Box and that the network connectivity is not blocked by firewall or other services on your host machine. Load the downloaded .ova file in Virtual Box, follow the 'Network setup' instructions from the Technical Documentation and access the OpenGeneMed system at 192.168.56.103.
The following documentation files are included in the virtual machine in the folder /opt/applications/opengenemed/docs, or may be downloaded separately for perusal prior to installation of the software.
OpenGeneMed Demo Walk-through
The distributed version of OpenGeneMed comes with a pre-populated database. It contains information from few mock patients, with simulated samples, treatment assignments, eligibility criteria, etc. This is done to allow first-time users to navigate a fully functional system and evaluate OpenGeneMed capabilities in its default configuration. It is important to clear the database to prepare the system for deployment of real case studies. To make this operation quick and easy, we provided a script that empties the appropriate tables and initializes the content of the database. More information about this can be found in the Technical Documentation.
This document presents some step-by-step screenshots about the usage of the OpenGeneMed system. The data and information shared about patients, genomic mutations and treatments are simulated and for illustration purpose only. For more details on the software architecture and implementation of OpenGeneMed, please refer to our publication: Palmisano, A., Zhao, Y., Li, M. C., Polley, E. C., & Simon, R. M. (2017). OpenGeneMed: a portable, flexible and customizable informatics hub for the coordination of next-generation sequencing studies in support of precision medicine trials. Briefings in bioinformatics, 18(5), 723-734.
Warning: This page describes the default configuration of the system and does not reflect changes that your system administrator may have implemented in your version of OpenGeneMed. Before submitting any inquiry to the authors of this document, please contact your system administrator to make sure your inquiry is not connected to changes they made to the default configuration.
Patients at different stages of registration, annotation, and treatment assignment process are presented to demonstrate fundamental steps in the management of a clinical research project.
Access OpenGeneMed with an Administrative account to be able to view all the different steps and roles.
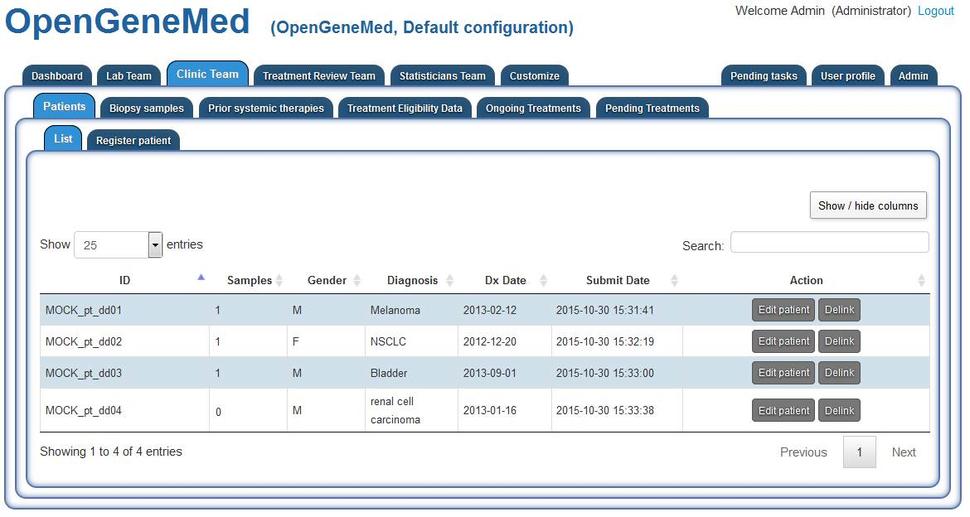
In the default configuration, login details of an Admin account are opengenemed_admin@example.com (password aaaAAA111). The current list of patients can be accessed from different tabs. Here we access it from the Clinic Team/Patients tab. Staff in different roles have different access to the list of patients.
Mock patient: General overview of a complete record
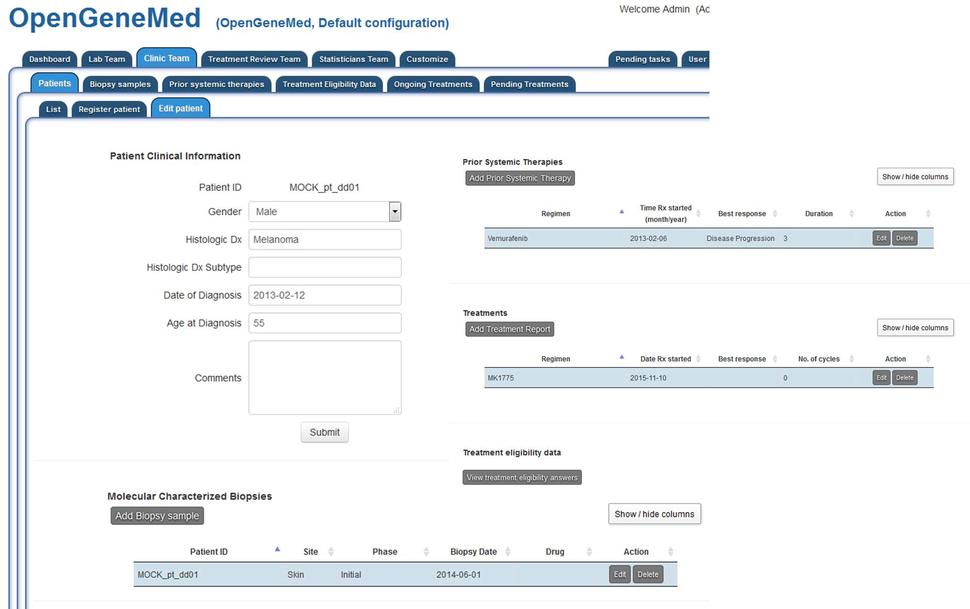
For patient "MOCK_pt_dd01", clicking the "Edit patient" from the table above will open the complete record for the patient. Here basic clinical information can be edited/deleted.
Mock Patient: Review variants, complete eligibility questions
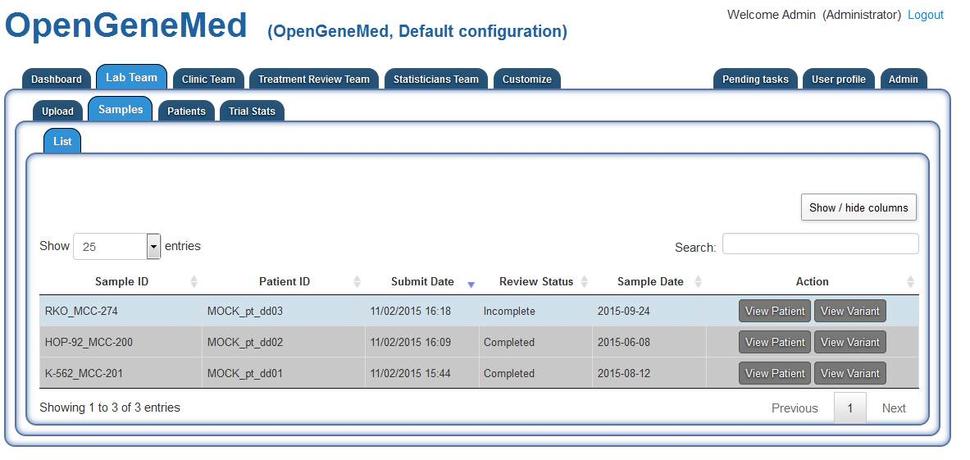
Accessing the "Lab Team" tab, we can see that patient "MOCK_pt_dd03" has an uploaded sample that still requires review (it is not greyed out). Grey rows indicate samples that have been reviewed and/or patients have already been assigned to treatment. The records remain in the table for future reference.
Clicking the "View variant" in the first row will open a more detailed view.
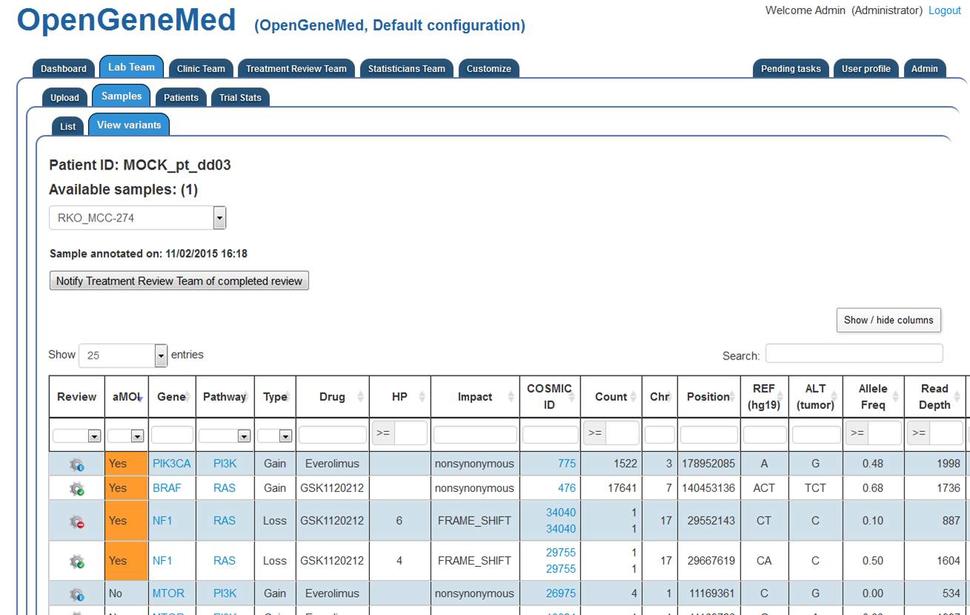
The variant mutation table is sortable and the user can hide/show columns of interest.
In particular, two of the four actionable mutations (the rows with aMOI "Yes") have a green icon (Review accepted status), one of them has a red icon (Review Rejected status) and one has a blue icon (Not yet reviewed).
By clicking the "Review" icon of the first variant (PKI3CA row), the user can see the specifics of the variant and decide on its review status. After reading all the information of the variant, the member of the Lab Team will make a judgment call (based on rules specified in the study protocol) and the variant can be marked as "Reviewed Approved". Once all the actionable mutations have been reviewed, the Lab Team member can notify the Treatment Review Team of the completed review by using the button right above the variant table.
Additional steps and functionalities are described in details in the OpenGeneMed user manual.