About OpenGeneMed
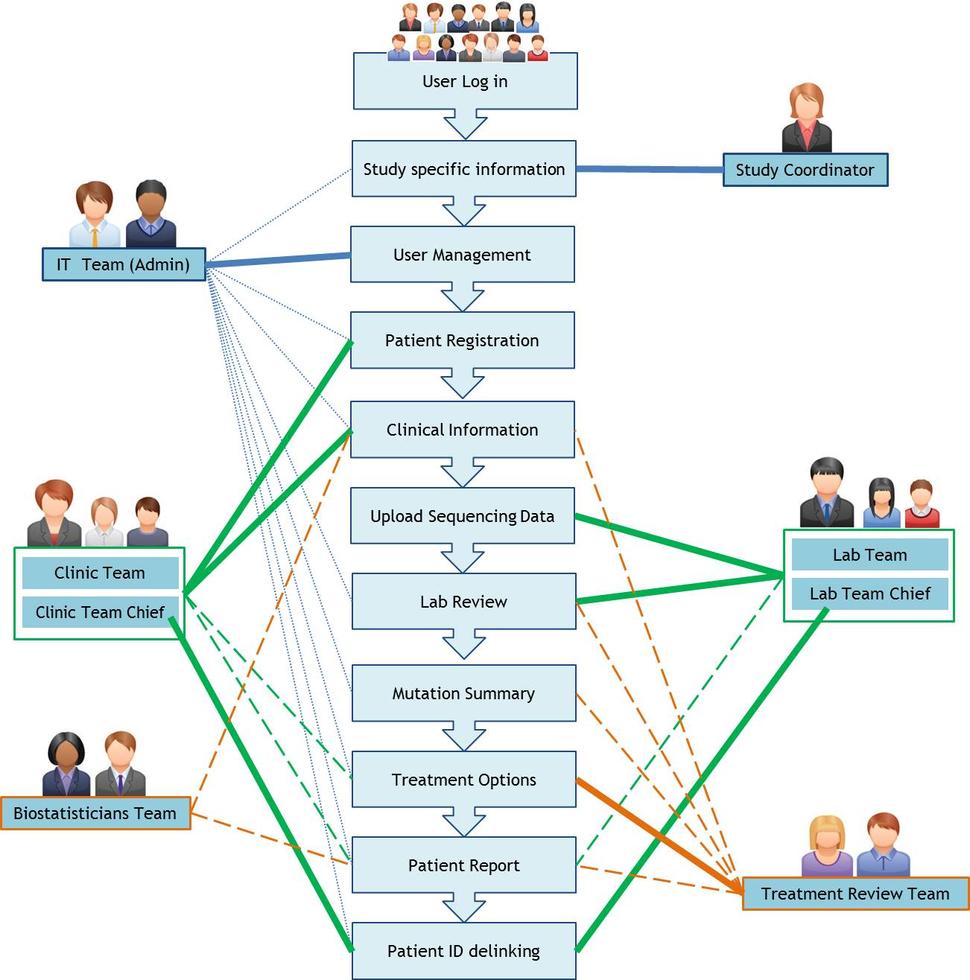
OpenGeneMed is an open and customizable version of the GeneMed system (Zhao et al, 2015). GeneMed is a web-based interface developed for the Molecular Profiling based Assignment of Cancer Therapy (NCI-MPACT) clinical trial, coordinated by the NIH. OpenGeneMed streamlines clinical trial management and it can be used by clinicians, lab personnel, statisticians, and other researchers as a communication hub. It automates the annotation of genomic variants identified by sequencing, classifies the actionable mutations according to customizable rules, and facilitates quality control by the molecular characterization lab in the review of variants. OpenGeneMed collects baseline information of patients to determine eligibility for the panel of treatment regimens available. The system generates patient reports containing detected genomic alterations along with summarized information that can be used for treatment assignment by a supervising treatment review team.
OpenGeneMed is distributed as a standalone virtual machine, ready for deployment and use from a web browser. OpenGeneMed code is modular, which allows for customization of existing features and addition of new modules to address the specific needs of different clinical trials and research teams. Examples of how to customize the code are provided in technical documentation that is distributed with the virtual machine. In summary, OpenGeneMed offers an initial set of features inspired by our experience with GeneMed, a system that has been proven to be an efficient and successful informatics hub for coordinating the application of next-generation sequencing in the NCI-MPACT trial.
Learn More
Palmisano A, Zhao Y, Li MC, Polley EC, Simon RM. OpenGeneMed: a portable, flexible and customizable informatics hub for the coordination of next-generation sequencing studies in support of precision medicine trials. Brief Bioinformatics. 2017;18(5):723-734. doi:10.1093/bib/bbw059 [PubMed Abstract]
Download OpenGeneMed
OpenGeneMed was developed by Biometric Research Team (BRP): Alida Palmisano, Ming-Chung Li, Eric Polley, Yingdong Zhao, Richard Simon. We thank Henry Rivera for a preliminary implementation of the OpenGeneMed.
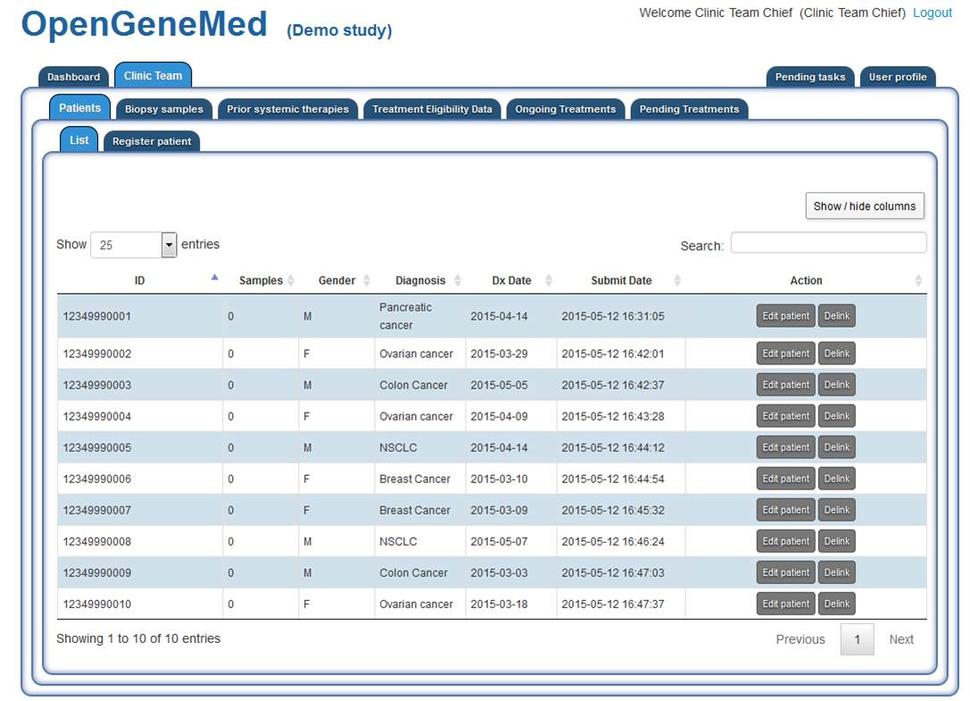
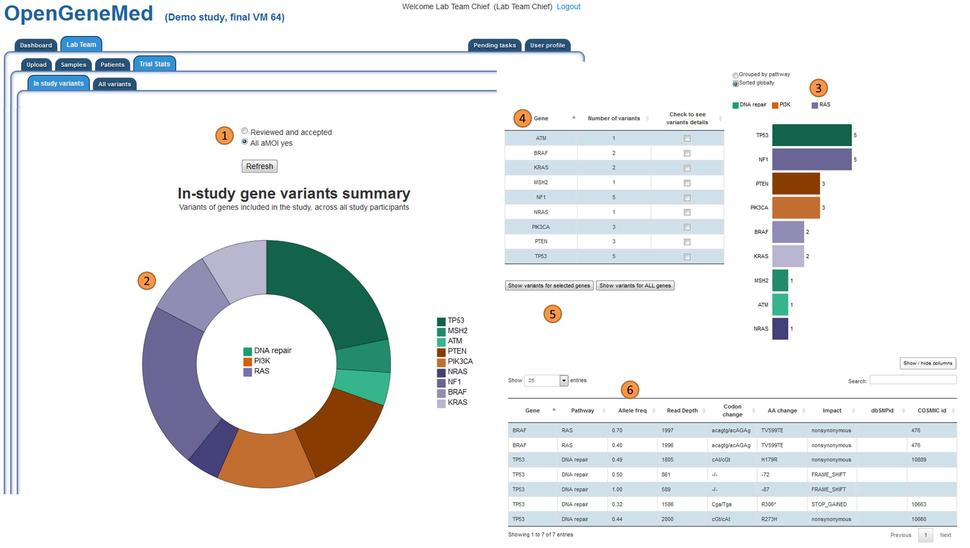
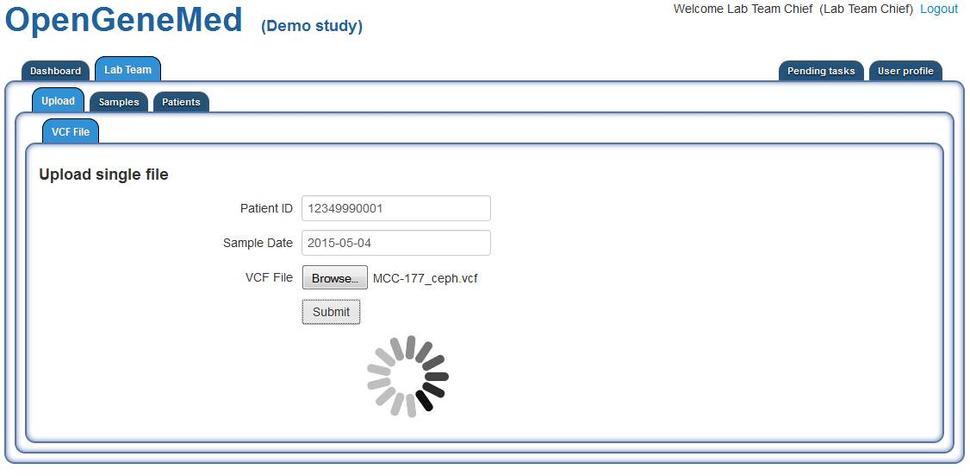
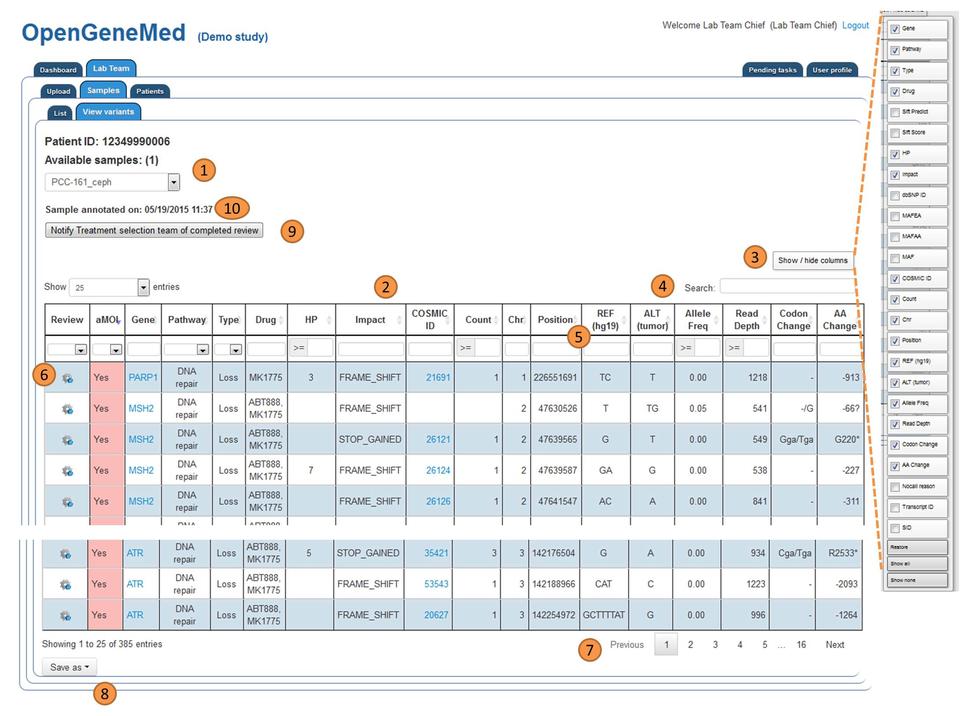
OpenGeneMed Screenshots
Please refer to the OpenGeneMed User Manual for more details and full description of all the software features.