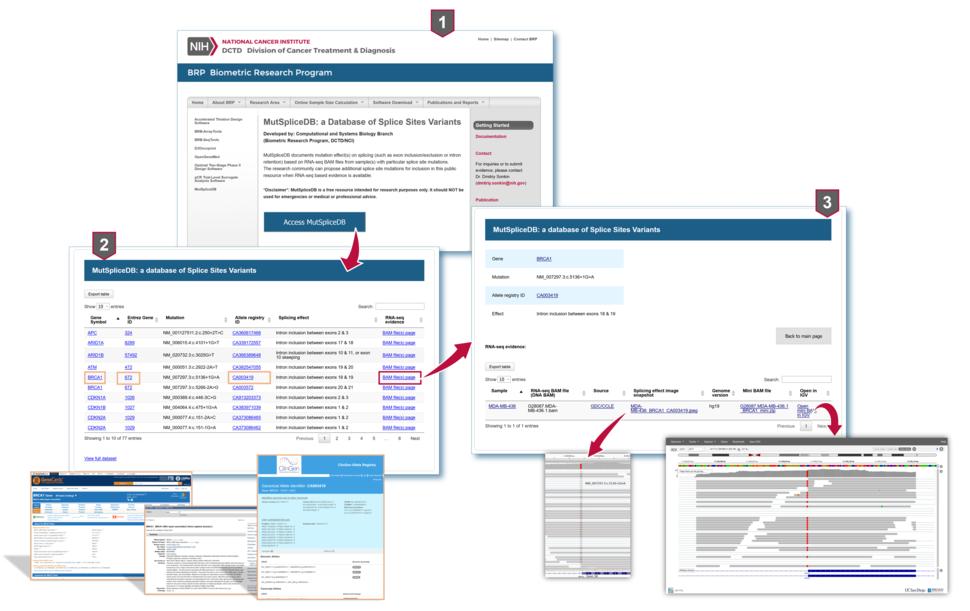
MutSpliceDB: A Database of Splice Sites Variants
MutSpliceDB documents mutation effect(s) on splicing (such as exon inclusion/exclusion or intron retention) based on RNA-seq BAM files from sample(s) with particular splice site mutations.
The research community can propose additional splice site mutations for inclusion in this public resource when RNA-seq based evidence is available.
Access MutSpliceDB
Inquiries and Evidence Submission
Email Dr. Dmitriy Sonkin (dmitriy.sonkin@nih.gov).
Publication
Palmisano A, Vural S, Zhao Y, Sonkin D. MutSpliceDB: A database of splice sites variants with RNA-seq based evidence on effects on splicing. Hum Mutatation. 2021;42(4):342-345. doi:10.1002/humu.24185 [PubMed Abstract]
Disclaimer
MutSpliceDB is a free resource developed by Computational and Systems Biology Branch (Biometric Research Program, DCTD/NCI) and it is intended for research purposes only. It should NOT be used for emergencies or medical or professional advice.
About MutSpliceDB
Splice site mutations are one of the well-known classes of genetic alterations playing an important role in biology. In cancer, splice sites are most frequently observed as inactivating alterations in tumor suppressor genes (for example, TP53 or RB1) and to a lesser degree as activating alterations in oncogenes (for example MET). Splice site mutations may lead to alterations in mRNA transcripts, causing for example exon(s) inclusion/exclusion or, intron retention. Interpreting the consequences of a specific splice site mutation is not straightforward, especially if the mutation is located outside of the canonical splice sites. Accurate interpretation of the impact a splice site mutation has can further our understanding of biology, influence patient treatment, and, in case of germline splice site mutations, may also have relevance to familial disease predisposition.
To facilitate the interpretation of splice site mutation effects, we developed MutSpliceDB: a database of splice sites variants, documenting mutation effect(s) on splicing based on RNA-seq BAM files from sample(s) with particular splice site mutations.
For each splice site mutation, the resource contains the following information:
- gene symbol;
- Entrez gene ID;
- HGVS compliant transcript based variant notation;
- allele registry ID;
- description of the splicing effect;
- sample name;
- sample source;
- name of RNA-seq BAM file;
- splicing effect image snapshot;
- mini BAM file with reads only for relevant gene (if there is no restrictions on nucleotide level data distribution);
- if the RNA-seq BAM file does not contain reads with splice site mutation (e.g., due to exon skipping), the name of BAM file with DNA sequencing data.
All entries in MutSpliceDB are based on publicly available RNA-seq BAM files. The initial release of MutSpliceDB (2019) contained detailed information for a subset of splice site mutations derived from publicly available RNA-seq data from Cancer Cell Lines Encyclopedia (CCLE) and The Cancer Genome Atlas (TCGA). We add information for more splice site mutations as soon as the necessary evidence becomes available.
How to Submit an Entry
MutSpliceDB is open for submissions from the molecular genetics community. Requests to add entry to MutSpliceDB should be addressed to Dr. Dmitriy Sonkin (dmitriy.sonkin@nih.gov) and should contain the following:
- All the splice site mutation information listed in the About MutSpliceDB section above,
- Image snapshots, and
- Mini BAM files (if there is no restrictions on nucleotide level data distribution) obtained as explained below.
Image Snapshot Requirements
Image snapshot files should show the splicing effect of the mutations and contain the following information:
- Gene Symbol,
- Relevant exon numbers and HGVS nomenclature compliant transcript based variant notation, and
- MANE Select/Plus transcript ID, if possible.
Image snapshot filenames should have the following structure: SampleName_GeneSymbol_AlleleRegistryID.jpeg.
For example, an image showing the splicing effects of TP53 mutation (NM_000546.5:c.375+5G>A) with Allele Registry ID CA645589233 in cell line PK-45H should have the following name: PK-45H_TP53_CA645589233.jpeg. Allele Registry ID for a variant can be found or generated using the ClinGen Allele Registry.
Mini BAM File Requirements
Mini RNA-seq BAM filenames should have the following structure: RNAseq BamFileName_GeneSymbol_mini.bam.
For example, mini BAM file for cell line PK-45H with TP53 mutation should have the following name: G27478.PK-45H.2_TP53_mini.bam. In this case G27478.PK-45H.2 is taken from the CCLE RNA-seq BAM file name G27478.PK-45H.2.bam.
To create the mini BAM files using Samtools, follow the steps below:
- samtools view RNA-seq.bam chr:start-end -b > mini.bam
- samtools index mini.bam
RNA-seq.bam file should be sorted and indexed. The instructions above create a sorted mini bam file (mini.bam) and the corresponding index file (mini.bam.bai). In Samtools view command 'chr' should be replaced with chromosome number, 'start' should be replaced with genomic position 100 bp before the start of first coding exon, and 'end' should be replaced with genomic position 100 bp after the end of last coding exon. Select the first and last coding exons in a way that covers all existing gene isoforms.