Overview
The PDMR’s primary source of tumor material for model development are two NCI-sponsored protocols:
- NCI Tissue Procurement Protocol (clincialtrials.gov: NCT00900198)
- CIRB Tissue Procurement Protocol 9846.
Specimen collection types:
- Fresh tissue
- Viably cryopreserved patient tumor tissue
- Material from rapid autopsies
- Previously-derived PDXs from outside groups.
All specimens have been collected under IRB-approved protocols and released through MTAs to allow use and distribution through the PDMR.
We continue to seek external site (profit and non-profit) contributors to provide previously-derived models or patient material to the PDMR for model generation and release to the research community. All models received by the PDMR go through the same QC procedures that are applied to internally-derived models.
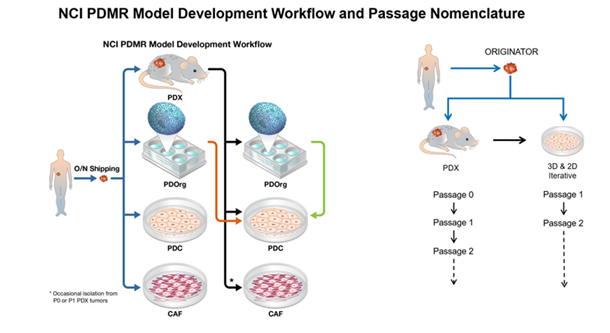
NCI Patient-Derived Model Development and Passage Nomenclature
Patient-derived xenograft (PDX), organoid (PDOrg), cell line (PDC), and cancer-associated fibroblast (CAF) models can be developed directly from patient material; PDOrgs, PDCs, and CAFs can also be developed from a PDX model; and PDCs can be developed from PDOrgs (see figure).
Originator material is the specimen obtained directly from the cancer patient.
- PDX model nomenclature begins with passage 0 (P0) which represents tumor material obtained from a mouse that was implanted with Originator material and allowed to grow.
- PDOrg and PDC models begin at passage 1 (P1) representing the first time that patient-derived tissue has been cultured in vitro.
PDX Models
A PDX is defined as any tumor model generated by implantation of fresh human tissue (tumor, enriched CTCs, etc.) into immunodeficient mice. The PDMR host mouse model used is NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG).
- The PDMR expands its models primarily by subcutaneous fragment engraftment to maintain as much tumor heterogeneity as possible.
- Distribution lots are early passage, generally P1-P4, which enables variations in histology and low allele frequency genetic variants to be observed in lineages within a model.
- These same types of variations can be expected in PDX models expanded by the recipients of distributed fragments from the repository.
- While PDX models generally demonstrate a high degree of stability, the PDMR has observed some models that continue to evolve; this caveat should be considered by any investigator requesting models for their research aims.
PDOrg Models
The PDMR generates PDOrg in vitro models from patient material and then PDXs (in that order of priority) to try to maximize model heterogeneity.
PDOrg cultures use specialized defined media with growth supported in a basement membrane dome. PDMR SOPs should be used to ensure culture success — these models should not be treated like traditional in vitro cultures (e.g., HeLa, MCF7).
The PDMR has noted different morphological patterns of growth among PDOrgs: organoids, loose aggregate clusters, and single cell/small clusters.
PDC Models
The PDMR generates PDC in vitro models from patient material, PDX, and patient/PDX-derived organoids (PDOrg) (in that order of priority) to try to maximize model heterogeneity.
PDC cultures use defined media and grow with a variety of growth characteristics. PDMR SOPs should be used to ensure culture success — these models should not be treated like traditional in vitro cultures (e.g., HeLa, MCF7).
CAF Models
CAFs are primarily generated from patient material, though occasionally they can be recovered from a P0/P1 PDX.
CAF cultures use defined media and are grown on Matrigel-coated plates.
PDMR Stringent Quality Control Criteria
Models are distributed only after the PDMR has performed multiple QC steps. As a general reminder, distributed material (cryopreserved PDX fragments, DNA vials, RNA vials, and fresh-frozen fragment vials) contain a mixture of human tumor and mouse stroma.
- Identifiler Short Tandem Repeat (STR) profiles are provided at the patient-level in the PDMR database for model validation.
- STR profiles are generated for all distribution lot material and are reported in toto for a model. For example, if a model has PDX and PDC material, all STR profile variants will be reported.
- The STR profile for any model derivative (PDX, PDC, CAF, PDOrg) is provided for reference.
- These profiles should be used to validate the model at multiple points during the experimental process.
- Human DNA content is analyzed by qRT-PCR to verify model origin.
- Pathology and/or IHC markers are confirmed relative to the patient diagnosis with all tumor models by a clinical pathologist.
- Concordance of STR profile and whole exome sequencing (WES) across all characterized model data.
PDX Model-Specific Quality Control
- Derived PDX models are confirmed by histopathology to match the patient diagnosis or pathology of patient originator material.
- Identifiler (STR profile) is performed on a fragment from every PDX to confirm lineage identity.
- A fragment from every PDX contributing to a distribution lot undergoes histopathology assessment to confirm diagnosis.
- PDX tumors that lose histological/morphological characteristics (e.g., de-differentiation) are screened with an IHC panel including markers for human content (Ku80 or hMito), CD45, EBV, cytokeratin, and/or vimentin to confirm the presence of human tumor material (aka high-grade tumor) versus human lymphoma or murine tumor out-growth.
- Additional markers may be used for certain cancer sub-types.
- Coming Soon: Breast cancer hormone receptor IHC validation
- A fragment from every PDX contributing to a distribution lot undergoes PCR assessment for human:mouse DNA ratio. Several models have stably low tumor content (20%-30%) relative to mouse stroma.
- PDX tumors that lose histological/morphological characteristics (e.g., de-differentiation) are screened by PCR to confirm murine tumor out-growth has not occurred.
- Each model has been successfully regrown from viably cryopreserved fragment.
- 3-5 fragments are implanted and monitored for 300 days for regrowth since some models can take up to 200-250 days to regrow to 1000 mm3 from a cryo-preserved sample.
- If the initial set of implants fails to grow, 3-5 additional fragments from a different cryo-lot are implanted and assessed.
- To date, very few models have failed this QC step, though some models have a higher percentage take-rate from frozen stock than others.
- Histopathology is also reconfirmed for all PDX regrown from viably cryopreserved fragments.
PDOrg Models
- Verified to be 99.9% human tumor culture, non-clonal (multiple cell types or non-cloned lineages), 99.9% fibroblast free (FACs analysis).
- PDOrg-derived xenografts confirmed by histopathology to match the patient diagnosis or pathology of patient originator material.
- Proven to grow as a cell-line derived xenograft (CLX) in NSG host mice. CLXs are characterized by histopathology for human tumor/diagnosis sub-type confirmation but are not further analyzed.
- Guaranteed for at least 10 passages from the distribution lot without outgrowth of fibroblasts when following PDMR SOPs.
- May or may not be able to form spheroids in a fully-defined, serum- and feeder-free medium
PDC Models
- Verified to be 99.9% human tumor culture, non-clonal (multiple cell types or non-cloned lineages), 99.9% fibroblast free (FACs analysis).
- PDC-derived xenografts confirmed by histopathology to match the patient diagnosis or pathology of patient originator material.
- Proven to grow as a cell-line derived xenograft (CLX) in NSG host mice. CLXs are characterized by histopathology for human tumor/diagnosis sub-type confirmation but are not further analyzed.
- Guaranteed for at least 20 passages from the distribution lot without outgrowth of fibroblasts when following PDMR SOPs.
- Tested for growth as spheroids in fully-defined, serum- and feeder-free medium and tested for clonal growth in soft agar.
CAF Models
- Verified to be 99.9% human fibroblast culture and 99.9% human tumor cell free by FACS analysis.
- Proven to be non-tumorigenic following subcutaneous implantation into NSG mice and have been characterized as fibroblasts by at least one of the following methods: immunohistochemistry, qRT-PCR, or FACs.
- It is important to note these are non-transformed cells. CAFs have a finite lifespan in vitro.
- CAFs are guaranteed for experimental use for up to 3 passages when maintained on Matrigel-coated surface in the recommended defined media.
- Additional population doublings and subcultures are possible, but overall fitness of culture may deteriorate with subsequent passages.
- Not sequenced for the Repository, though they may be used as a germline surrogate for the model.
Available NextGen Sequencing (NGS) Data
- Whole-exome sequencing (WES); FASTQ and vcf
- Consensus WES (variants present in all sequenced PDX models); MAF and vcf
- RNASeq; FASTQ and TPM
- Germline sequence (when available); FASTQ and vcf
- Cancer-associated genes; table of reported variants from WES
NGS Generation
- The pathology, WES, RNASeq, etc. provided for each model in the PDMR database are derived from representative samples of that model. Recipients should characterize the PDX models during expansion for their experiments to ensure they remain consistent over time.
- Murine reads are bioinformatically removed from posted WES and RNASeq files in the PDMR database as per the posted PDMR Genomics SOPs; however, some murine contribution from the NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) host, the reference sequence, for which can be found on the SOP page, will likely remain.
- Patient germline sequence has been included at the patient-level in the database, and somatic calls have been made, for a limited set of models. These were generated from either a peripheral mononuclear blood fraction or purified cancer-associated fibroblasts (CAFs).
- For all other models, somatic mutations could not be removed from the model sequences.
File Naming Structure
- Sequence file names include the version of the pipeline used for sequencing.
- The most recent pipeline version of the sequence files are linked in the PDMR database.
- If you are obtaining data files through bulk downloads from the FTP site, be sure to use the most recent pipeline version for analysis. For example, an RNASeq file with version “v2.0” should be used instead of a file with “v1.2” for the same PDX.
- NOTE: the README file lists previously posted sequence files that should be removed from any subsequent analysis.
Limitations and Caveats of PDX Models
- Many PDX models grow slowly, both when coming out of the freezer, as well as between passages. For some models, you should expect initial implants to take as long as 200 days before tumor is of sufficient size for passage. Representative growth curves for each PDX model are available in the PDMR database in the specimen details.
- Several models have stably low human tumor content (e.g., 20%-30%). The remaining content would be murine stroma. Pathology Reports are available in the PDMR database for quick tumor/stromal content review.
- The PDMR serially passages tumor fragments (not cell slurries) directly from mouse to mouse so that every PDX lineage can be traced back to the patient tumor. Since intra-tumor heterogeneity can be observed in the models, we cannot guarantee which fragment a requestor will receive. For instance, there may be variation in differentiation level of the tumor or for a stomach adenocarcinoma in the signet ring cell content.