Translational Research Program in Cancer Differences across Populations
Fred Hutchinson Cancer Center
Principal Investigator(s)
Ulrike Peters, Ph.D., M.P.H.
Professor, Associate Director, Endowed Chair
Public Health Sciences Division
Fred Hutchinson Cancer Center
1100 Fairview Ave N, M4-B402
Seattle, Washington, 98109
206-667-2450
Christopher Li, M.D., Ph.D.
Professor, Public Health Sciences Division
Vice President, Faculty Affairs and Diversity
Fred Hutchinson Cancer Center
1100 Fairview Ave N, M4-C308
Seattle, Washington, 98109
206-667-5948
Timothy Thomas, M.D.
Physician Researcher, Research Services Department
Division of Community Health Services
Alaska Native Tribal Health Consortium
4000 Ambassador Dr,
Anchorage, Alaska, 99508
907-563-2662
Overview
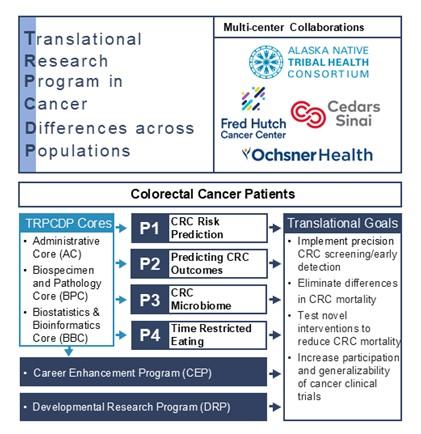
The Translational Research Program in Cancer Differences across Populations (TRPCDP) was specifically developed to address differences in cancer incidence and mortality in the United States, with a particular focus on colorectal cancer (CRC).
During the P20 phase, we assembled a comprehensive biorepository containing various biospecimen types and detailed clinical data from CRC patient populations with varying incidence and mortality rates. Additionally, our established leadership in genetic epidemiology has provided access to the world's largest CRC germline genetic dataset spanning multiple genetic ancestries.
Building upon these unique resources, our P50 program will implement four translational research projects supported by three essential cores. To achieve our overarching goal of reducing CRC incidence and mortality across populations, we will address critical knowledge gaps by pursuing the following specific aims.
Aim 1: Improve risk-stratified screening and early detection of CRC across population groups. We will develop clinically actionable risk prediction models that perform consistently across genetic ancestral groups and inform the selection of primary prevention strategies (SPORE Project 1).
Aim 2: Reduce differences in CRC-specific mortality. We will discover and validate novel molecular and biological markers associated with CRC mortality and treatment response to guide surveillance strategies and treatment selection for CRC survivors (SPORE Projects 2 and 3).
Aim 3: Discover novel therapeutic targets and test clinical interventions to reduce CRC mortality across different populations. We will elucidate the genetic, molecular, and microbial characteristics that distinguish CRC across different populations, leading to the development of targeted therapeutic strategies. We will evaluate the effectiveness of these novel interventions through rigorous clinical trials (SPORE Projects 1, 2, 3, and 4).
Project 1: Functional guided risk prediction for colorectal cancer across genetic ancestries
Project Co-Leaders
- Anshul Kundaje, Ph.D. (Stanford University) (Basic Co-Leader)
- Li Hsu, Ph.D. (FHCC) (Clinical/Applied Co-Leader)
- Ulrike Peters, Ph.D., M.P.H. (FHCC) (Clinical/Applied Co-Leader)
This project will develop genetic risk prediction models for colorectal cancer that perform equitably across genetic ancestries in the US.
Aim 1: Develop functional genomic-Informed Genetic Risk Prediction Models. In Aim 1a, we will create functional variant effect scores using comprehensive molecular data from normal colorectal tissue. This approach employs deep learning algorithms to predict how genetic variants influence gene expression and regulatory activity within colorectal tissue at high resolution. In Aim 1b, we will integrate these functional scores with large-scale genome-wide association study (GWAS) data to construct precise polygenic risk scores (PRS) that explicitly account for genetic ancestry in statistical modeling.
Aim 2: Evaluate Cost-Effectiveness of PRS-Guided Screening Strategies. Using microsimulation modeling, we will assess the cost-effectiveness of implementing PRS for risk stratification and determine optimal screening strategies across genetic ancestral populations in the United States. This analysis will examine the distribution of costs, benefits, and screening efficiency by genetic ancestry, comparing PRS-guided approaches with current screening guidelines while incorporating relevant environmental risk factors.
As PRS implementation in clinical care accelerates, this project has significant potential to advance CRC prevention through personalized interventions. By reducing the high CRC burden and differences in incidence and mortality across population groups, our approach will contribute to improved cancer prevention strategies.
Project 2: Discovery and validation of novel molecular and immune predictors of colorectal cancer mortality and response to treatment across patient population groups
Project Co-Leaders
- Li Li, Ph.D. (Ochsner Health) (Basic Co-Leader)
- Christopher Li, M.D., Ph.D. (FHCC) (Clinical/Applied Co-Leader)
- Marc Matrana, M.D. (Ochsner Health) (Clinical/Applied Co-Leader)
This project seeks to uncover new biomarkers in colorectal cancer (CRC) that predict patient outcomes and treatment responses across population groups.
Aim 2 aims to predict treatment responses: We will develop tools to forecast responses to standard CRC therapies across population groups, leveraging insights from Aim 1. We will also study cell patterns in tumors of 100 CRC patients treated with immune checkpoint therapy to predict treatment effectiveness.
Aim 1: Identify Spatially Resolved Prognostic Biomarkers. We will analyze CRC tumor tissues from 840 patients using advanced spatial profiling technology (PhenoCycler) to identify prognostic biomarkers based on the spatial organization and interactions of immune and cancer cells. This approach will enable prediction of CRC-specific mortality by leveraging the tumor microenvironment architecture.
Aim 2: Develop Treatment Response Prediction Models. We will develop tools to forecast responses to standard CRC therapies across population groups, leveraging insights from Aim 1. We will also study cell patterns in tumors of 100 CRC patients treated with immune checkpoint therapy to predict treatment effectiveness.
Aim 3: Integrate Multi-Modal Biomarker Data for Clinical Translation. We will combine findings from Aims 1 and 2 to develop computational models predicting three key outcomes: CRC mortality risk, response to standard treatments, and response to immunotherapy. These models will be validated with patient data to ensure accuracy and reliability.
This project aims to improve identifying high-risk CRC patients and personalize treatments, potentially enhancing monitoring and offering alternative therapies to those with increased rates of CRC-related mortality. New predictive tools will have a positive impact on reducing long-standing population differences in CRC outcomes by aiding clinicians in selecting optimal treatments. Additionally, insights into immune responses to CRC and treatments could advance future therapies.
Project 3: Population group differences in the intra-tumoral microbiome: Impact on colorectal cancer mortality and clinicopathologic correlates
Project Co-Leaders
- Meredith Hullar, Ph.D. (FHCC) (Basic Co-Leader)
- Amanda Phipps, Ph.D. (FHCC) (Clinical/Applied Co-Leader)
This project investigates the role of tumor-associated bacteria in colorectal cancer (CRC) across population groups to identify differences in bacterial composition and clinical associations.
Aim 1: Characterize Bacterial Distribution and Prognostic Associations. Will use digital droplet PCR assays to measure new types of bacteria in colorectal tumors (a) to look for differences in how these bacteria are distributed among CRC patients across four population groups and (b) to see if these bacteria are linked to a higher risk of deadly CRC overall and across population groups.
Aim 2: Analyze Microbiome Composition by Clinicopathologic Factors. Will study how the composition of the tumor-associated microbiome differs based on clinicopathologic factors that vary across population groups and affect prognosis (like where the tumor is located, age at CRC diagnosis, and tumor marker status).
Aim 3: Develop AI-Based Prognostic Models. We will apply artificial intelligence and machine learning algorithms to 16S rRNA sequencing data to identify specific microbial signatures in CRC tumors that predict CRC-related mortality. We will also examine how these prognostic signatures vary among different population groups.
This research may identify novel bacterial biomarkers for enhanced early detection strategies, particularly benefiting high-risk populations. For patients with established diagnoses, characterizing tumor-specific bacterial profiles could enable precision therapeutic approaches, including targeted antimicrobials, novel drug delivery systems, bacteriophage therapy, and personalized dietary interventions.
Project 4: Effect of Meal Timing during Cancer Treatment in Alaska Native Patients: A Randomized Clinical Trial
Project Co-Leaders
- Jane Figueiredo, Ph.D. (Cedars Sinai) (Basic Co-Leader)
- Timothy Thomas, M.D. (ANTHC) (Clinical/Applied Co-Leader)
Alaska Native (AN) people experience disproportionate burden from metabolic diseases and have the highest colorectal cancer (CRC) incidence and mortality rates globally. This proposed clinical trial builds upon the research infrastructure established through our P20 SPORE planning grant (P20CA252733) and our ongoing randomized clinical trial of time-restricted eating (TRE) in rectal cancer patients (CHRONO Trial, R01CA258222, NCT04722341). We aim to conduct a randomized Phase II clinical trial enrolling 100 AN rectal cancer patients receiving treatment at the AN Medical Center.
Aim 1: Evaluate TRE Impact on Treatment Outcomes and Patient Experience. We will determine the effects of TRE during neoadjuvant treatment on treatment response, treatment-related toxicities, quality of life, and adherence among AN patients with rectal cancer. Primary endpoints include pathological complete response rates, treatment-related adverse events, health-related quality of life measures, and TRE adherence rates.
Aim 2: Characterize TRE Effects on Metabolic and Cancer-Related Biomarkers. We will analyze blood-based biomarkers including IGF-1, IGFBP-3, insulin, glucose, C-reactive protein, and DNA damage markers in peripheral leukocytes to assess TRE's impact on metabolic function, inflammation, and genomic stability. Additionally, we will examine cellular and molecular changes in rectal tumor tissue specimens.
Aim 3: Compare Outcomes Across Population Groups. We will examine differences in clinical outcomes, adherence rates, and biomarker responses across five distinct population groups by comparing findings from this AN-focused study with data from different population groups enrolled in the parallel CHRONO Trial.
The proposed trial is innovative in testing a promising, accessible intervention in a population with a high burden of CRC and metabolic disease.
Administrative Core
Core Directors
- Ulrike Peters, Ph.D., M.P.H. (FHCC)
- Christopher Li, M.D., Ph.D. (FHCC)
- Timothy Thomas, M.D. (ANTHC)
The Administrative Core (AC) will provide overall leadership and administrative support, program management, integration with existing programs and shared resources, infrastructural support, communication within and across TRPCDP investigators and partnering institutions, and evaluation of all TRPCDP activities and components.
This Core has the following specific aims
- Aim 1. Leadership: To provide the organizational structure and scientific and administrative leadership needed for the successful execution and evaluation of key TRPCDP objectives.
- Aim 2. Administrative Management: To provide administrative management of all SPORE components including fiscal oversight, regulatory compliance, SPORE leadership succession plans, communication, data management, and engagement with NCI.
- Aim 3. Integration of the P50 Program with the host institutions: To coordinate TRPCDP activities with various projects, programs, and shared resources at partnering institutions such that integration of these existing center resources will successfully support the TRCPDP.
- Aim 4. Planning and evaluation of SPORE activities: To plan SPORE activities, assist with and prepare progress reports, organize regular Advisory Board meetings, and support evaluations conducted by our Advisory Boards.
- Aim 5. Sustain collaborations: To develop and support multi-center intra- and inter-SPORE interactions and initiate and sustain collaborations with key external partners.
Biospecimen and Pathology Core
Core Directors
- Amanda Koehne, D.V.M., Ph.D. (FHCC)
- Cecilia Yeung, M.D. (FHCC)
The Biospecimen and Pathology Core (BPC) has set up a robust system for collecting, processing, and providing access to biospecimens across institutions. This resource meets the critical need for biospecimens from colorectal cancer (CRC) patients. The BPC offers advanced laboratory technologies and specializes in tissue-based assays and biomarker analyses.
This Core has the following specific aims
- Aim 1 is to establish a high-quality CRC biospecimen repository from CRC patients, including blood, frozen and preserved tumor and normal tissues, saliva, urine, and stool, all meticulously annotated.
- Aim 2 involves processing, evaluating, and storing biospecimens, with services such as pathology review, H&E staining, tumor microarray development, digital imaging, DNA extraction, and single cell preparation. These biospecimens will support SPORE, CEP, DRP projects, and external collaborations.
- Aim 3 focuses on comprehensive data collection, harmonizing demographic, lifestyle, clinical, population level health factor data, and outcomes data. Medical chart abstractions for CRC patients and data harmonization from epidemiological studies maximize research value.
- Aim 4 through the TRPCDP Biospecimen Utilization Committee, prioritizes access based on merit of the research project, optimizing research across SPORE, CEP, DRP projects, and external collaborations.
- Aim 5 expands technological capabilities, leveraging Institutional Shared Resources to offer advanced technologies supporting innovative research across SPORE, CEP, DRP, and external partnerships.
Biostatistics and Bioinformatics Core
Core Directors
- Li Hsu, Ph.D. (FHCC)
- Jeroen Huyghe, Ph.D. (FHCC)
The Biostatistics and Bioinformatics Core (BBC) is pivotal in uncovering population differences in CRC incidence and mortality by employing systematic and rigorous approaches. By conducting research across all phases—from standardized study designs and data collection methods to sophisticated analyses—the BBC aims to identify molecular and genomic features linked to colorectal cancer (CRC) risk and mortality. This knowledge will inform improved prevention, diagnosis, and treatment strategies.
Aim 1 focuses on providing statistical and bioinformatic support in risk prediction, observational studies, survivorship, treatment response, and clinical trials. Utilizing advanced omics data like single cell ATACseq and RNAseq, genome-wide genetic sequencing, digital spatial single cell profiling, microbiome profiling, and blood-based biomarkers, the BBC collaborates on study design, ensures data quality, conducts analyses, and interprets results biologically using bioinformatics tools. The BBC also supports manuscript preparation and result dissemination across population groups to ensure accurate and valid data presentation.
Aim 2 involves monitoring methodological advancements and evaluating state-of-the-art statistical methods and computational tools to handle complex data from SPORE projects. The BBC adapts existing methods or develops new ones as needed to propel SPORE research forward.
Developmental Research Program (DRP)
Program Directors
- William Grady, M.D. (FHCC)
- Li Li, Ph.D. (Ochsner Health)
The Developmental Research Program (DRP) provides key support to develop novel translational early-phase projects that have the potential to substantially impact our understanding of differences in cancer incidence and mortality rates across population groups and ultimately, will evolve into full projects.
Our DRP is designed to achieve several specific aims: First, it supports innovative early-stage cancer research across translational areas such as molecular biology, epidemiology (including primary and secondary prevention), risk prediction, early detection, treatment, and survivorship. Second, it encourages high-risk, high-reward pilot projects to maintain the leading edge of our SPORE research efforts. Third, the program actively involves investigators in leading these early-stage research initiatives. Fourth, it provides essential funding, infrastructure, and mentorship to support the feasibility and success of DRP projects. Fifth, the DRP assists in advancing the projects to becoming full research endeavors.
Career Enhancement Program (CEP)
Program Directors
- Christopher Li, M.D., Ph.D. (FHCC)
- Diana Redwood, Ph.D. (ANTHC)
The Career Enhancement Program (CEP) identifies and supports promising junior investigators, and those new to the field of translational research aimed at meaningfully reducing long-standing differences in cancer incidence and mortality across population groups. Recruitment of candidates will be conducted across all partner sites, and tailored career enhancement programs will be developed to meet the needs of each awardee. The CEP will bring together junior, mid-level, and senior investigators by providing funding and scientific support for innovative projects that study differences in cancer incidence and mortality across population groups. By providing mentorship and finding sponsors within their institutions, the CEP will help participants develop their careers in this specialized field. The CEP will provide leadership training and actively support the hiring of new researchers focused on translational research on cancer risk and outcomes among population groups. Our goal is to strengthen our research community and make significant progress in reducing differences in cancer rates and outcomes.