Global Multiomics Study Reveals Key Lung Cancer Markers and Targets
- Posted:
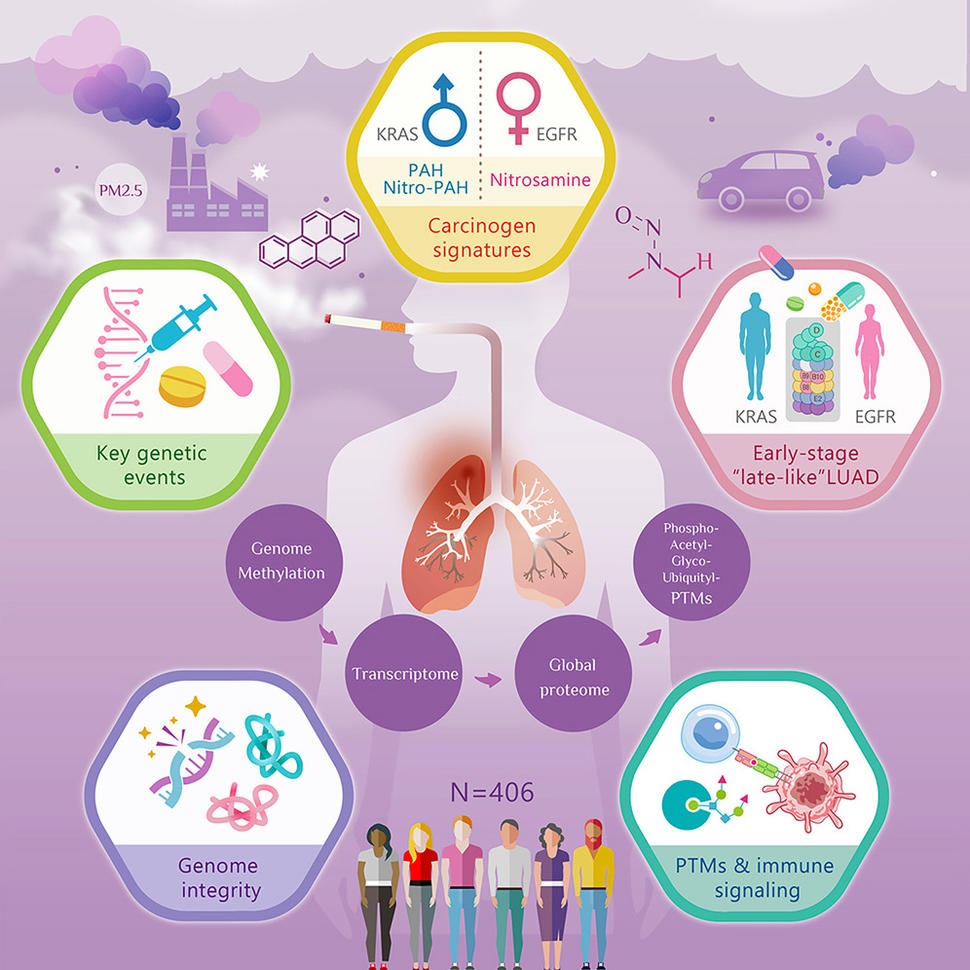
An infographic depicting the multi-omic analysis of lung adenocarcinoma (LUAD) in a cohort of N=406 patients
Credit: Cancer Cell
A new study, published in Cancer Cell, is the result of an international collaborative effort to examine the effects of ethnicity, smoking, environmental exposures, and sex on lung adenocarcinoma (LUAD) via large-scale multiomics analysis.
Researchers from the U.S. National Cancer Institute's Clinical Proteomic Tumor Analysis Consortium (CPTAC) and the International Cancer Proteogenome Consortium (ICPC) Taiwan team (Academia Sinica, Taiwan Cancer Moonshot Program) analyzed 406 tumors and 388 matched normal adjacent tissues from North American, Eastern European, and Asian populations representing a variety of smoking histories.
Their findings, including oncogenic mechanisms, prognostic markers, sex-specific carcinogen effects, and ‘‘late-like’’ traits in early-stage tumors, represent a marked step forward in our understanding of cancer biology.
"This study is the largest proteogenomics study presented to date in any cancer and the product of direct collaborative work between CPTAC and the ICPC. That combination of unprecedented scale and demographic diversity enabled a highly nuanced exploration of lung adenocarcinoma,” said Michael Gillette, PhD, co-senior author from the Broad Institute.
New Insights into Tumor Biology and Prognosis
Previous studies have demonstrated that standard measures of chromosomal instability (e.g. weighted genome instability index) fail to predict patient outcomes in LUAD. In response, this study presents a new metric coined Breakage Intensity Clustering (BIC), which classifies tumors by analyzing the clustering of DNA breakpoints.
BIC successfully stratified patients into three groups (contiguous, fragmented, and intense) with significantly different survival outcomes. Tumors classified as BIC-intense (poor prognosis group) showed amplifications of key oncogenes such as TERT, MYC, and NKX2-1. Furthermore, the protein IGF2BP3 was identified as both a robust proteomic biomarker for genomic fragmentation and as a predictor of immune checkpoint inhibitor response.
Proteomic Subtypes and the "Late-Like" Tumor Phenotype
Proteomic clustering analysis grouped tumors into three subtypes (C1, C2, and C3), with the C2 subtype being associated with a higher recurrence rate and shorter relapse-free survival (RFS). Tumors in the C2 cluster showed activation of pathways typically seen in advanced cancers and were highly enriched for TP53 mutations. This cluster also included a significant percentage (36.4%) of stage I tumors with very poor RFS, a characteristic described as "late-like" in a previous ICPC study.
Yu-Ju Chen, PhD, a co-corresponding author from Academia Sinica, Taiwan said, “This study integrates multiomic profiles from a large cohort spanning diverse ethnicities, sexes, and environmental exposures. The results confirm the cross-population relevance of a “late-like” LUAD subtype and support its potential as a foundation for the development of a universal in vitro diagnostic (IVD) tool.”
Decoding the Influence of Environmental Carcinogens
By analyzing mutational signatures, the study identified distinct tumor clusters associated with common carcinogens like polycyclic aromatic hydrocarbons (PAHs) and nitrosamines. Different carcinogens activated distinct cancer-promoting pathways; for instance, nitro-PAH/PAH signatures were linked to the AHR xenobiotic metabolism pathway, while the nitrosamine signature was associated with MAPK and ERBB signaling.
Notably, carcinogen exposure was found to affect not only the tumor but also the normal adjacent tissue by activating pathways related to inflammation and immune response contributing to a so-called ‘field of cancerization.’
Dr. Gillette commented, “While some key findings were specific to lung adenocarcinoma, others—such as the existence of early clinical stage tumors with late-like biological characteristics and outcomes, the prognostic significance of genomic fragmentation, and the influence of ubiquitous carcinogens such as nitrosamines—may have implications for other cancers.”
Therapeutic Opportunities and a Path Toward Precision Medicine
The study also identified drug targets and nominated potential drugs for different LUAD subtypes. Drug targets were prioritized if its protein, activating phosphorylation site, or other PTM site was overexpressed in a particular LUAD subtype and knocking down the gene was essential for the survival of corresponding cell lines.
Using this approach, the group identified numerous dependencies, including the splicing factor SF3B, the kinase MET, and the protein transporter XPO1. The identified targets were classified into five tiers based on their actionability, ranging from those targeted by approved drugs to potential candidates for novel therapies like antibody-drug conjugates or CAR T-cells.
"Our proteogenomic atlas of lung adenocarcinoma offers a critical new blueprint for precision medicine—uncovering actionable vulnerabilities and revealing previously unappreciated biology underlying tumor behavior,” said Shankha Satpathy, PhD, co-first author of the study from the Broad Institute. “Large-scale collaborations such as this not only bring exceptional talent together but also enable access to genetically and geographically diverse sample sets, making this study truly reflective of an LUAD that remains one of the leading causes of cancer-related deaths worldwide.”
View the data from this study in the Proteomic Data Commons.