Proteomics Provides Insights Into DNA-PK Inhibitor Mechanism in Patient Samples
Targeted proteomic analysis of patient samples as part of a Phase I/IIa clinical trial (NCT03907969) has yielded insights into how AZD7648, a DNA-dependent protein kinase (DNA-PK) inhibitor, alters DNA repair signaling. Using CPTAC-developed targeted proteomic assays [1, 2, 3], researchers from the Paulovich CLIA-CAP Lab at Fred Hutchinson Cancer Center were able to observe upregulation of proteins indicative of drug activity. Their findings appear in the British Journal of Cancer.
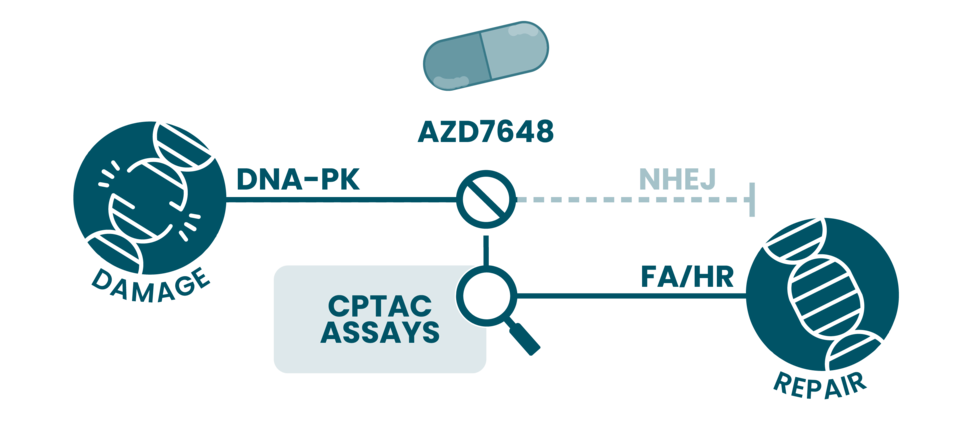
AZD7648 is a selective inhibitor of DNA-PK, whose upregulation is associated with poor prognosis and resistance to DNA-damaging therapies across multiple tumor types. The trial, designed to assess safety and efficacy of AZD7648, enrolled 30 patients with advanced malignancies who had progressed on prior treatments. Patients received AZD7648 either alone (n=14) or AZD7648 in combination with pegylated liposomal doxorubicin (n=16).
To evaluate pharmacodynamic responses, CPTAC investigators from the Paulovich CLIA-Cap Lab at Fred Hutch used targeted immuno-MRM mass spectrometry to peripheral blood mononuclear cells (PBMCs) collected during treatment. As a reference for DNA-PK inhibition, PBMCs from healthy volunteers were treated ex vivo with AZD7648, generating a proteomic signature characterized by increased abundance of proteins involved in Fanconi anemia and homologous recombination repair pathways, consistent with compensatory activation of alternative DNA repair mechanisms.
Patient PBMC samples were then compared against this ex vivo benchmark. No consistent modulation of DNA damage response proteins was observed at dose levels up to 40 mg. However, one patient treated with AZD7648 at 80 mg twice daily exhibited a proteomic profile closely resembling the ex vivo DNA-PK inhibition signature, suggesting biological target engagement at higher drug exposure.
While additional data are needed, these results demonstrate how CPTAC’s targeted proteomic assays can be applied in a CLIA-Cap environment to clinical trial samples to assess pharmacodynamic effects and target engagement. The study highlights the potential of targeted proteomics to support dose selection and biomarker strategies in the development of DNA repair inhibitors.